Learn with us
Learn more about immune biology, repertoire sequencing, and iRepertoire’s solutions for characterizing immune diversity.
Click a topic to see what articles are available:
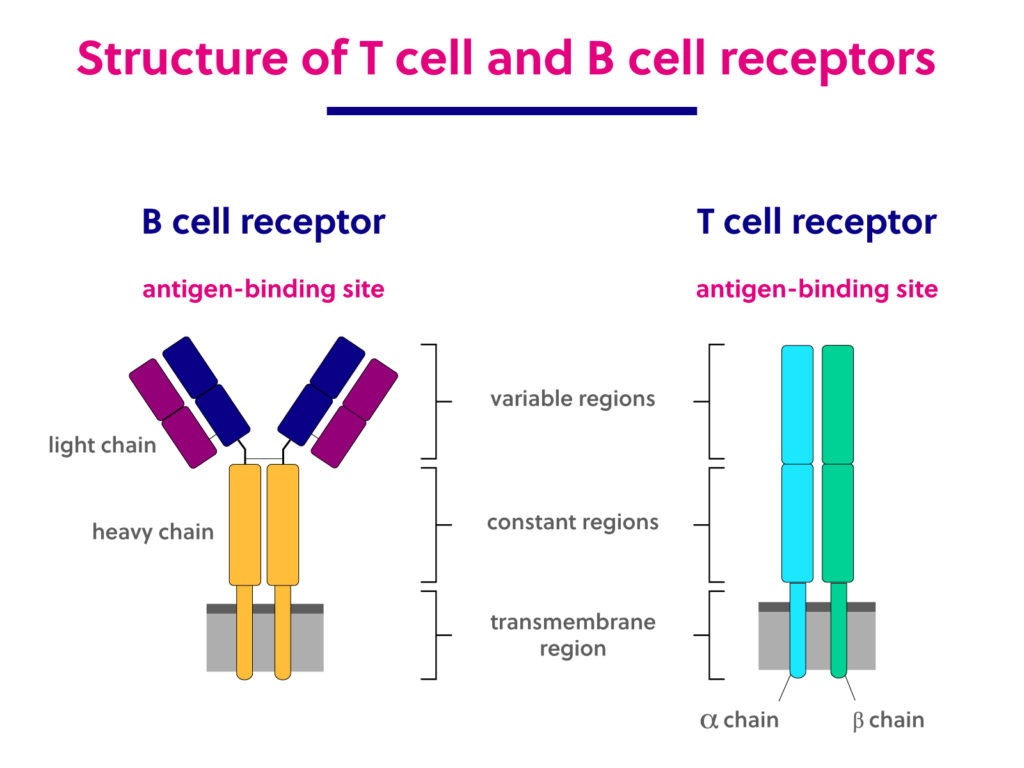
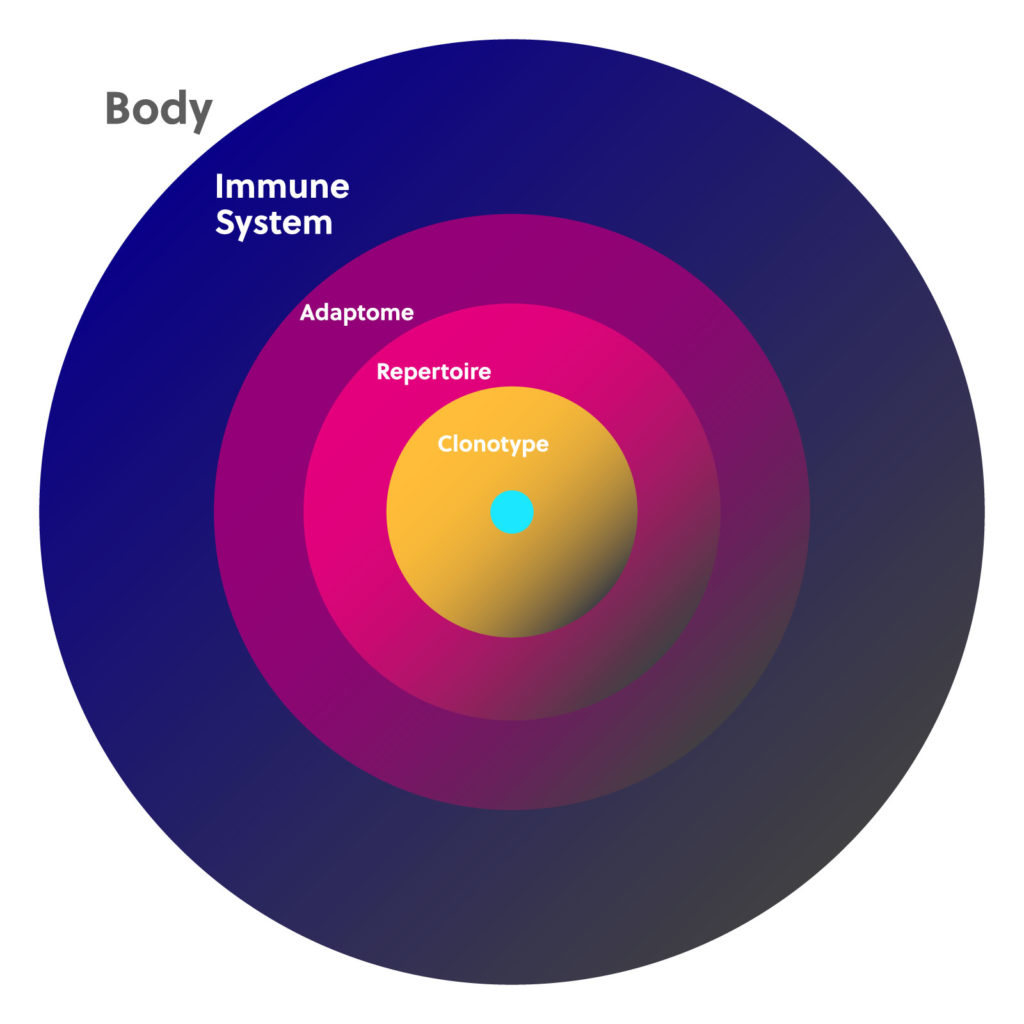
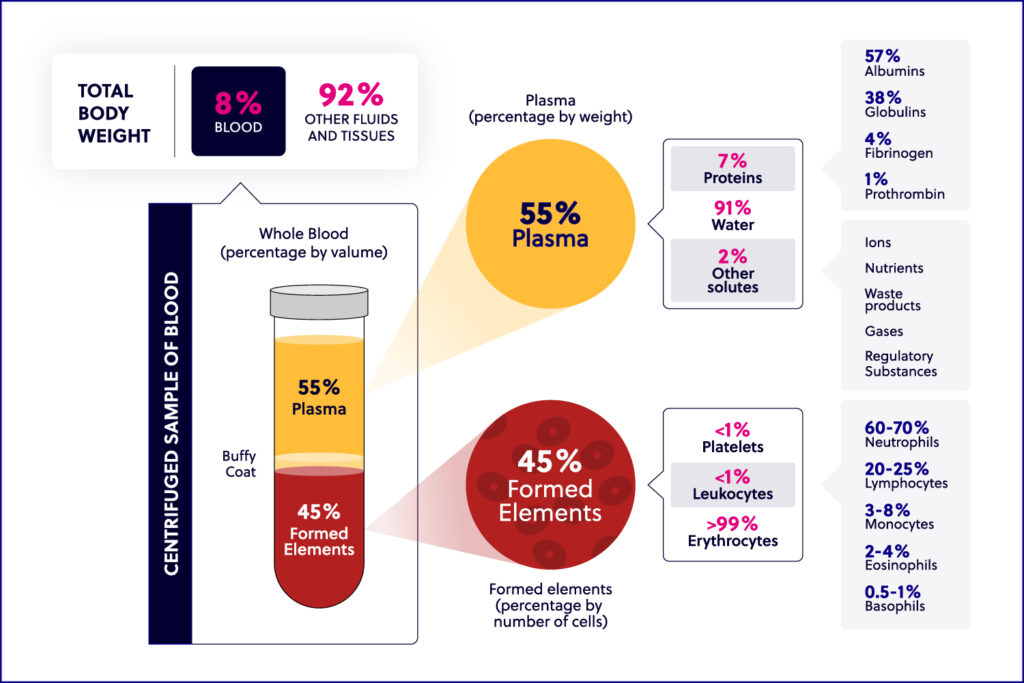
Immunology basics
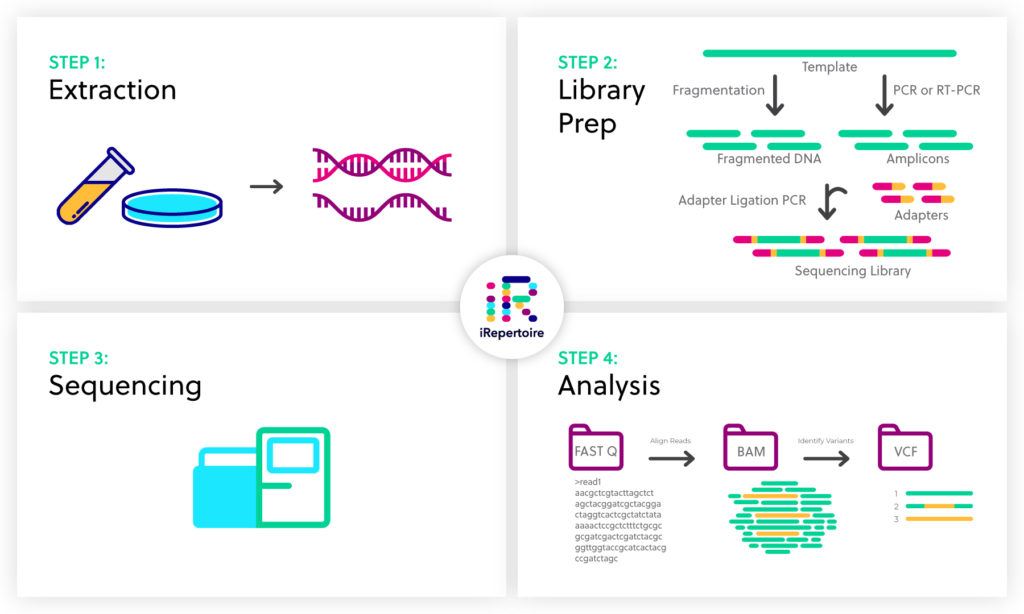
NGS basics
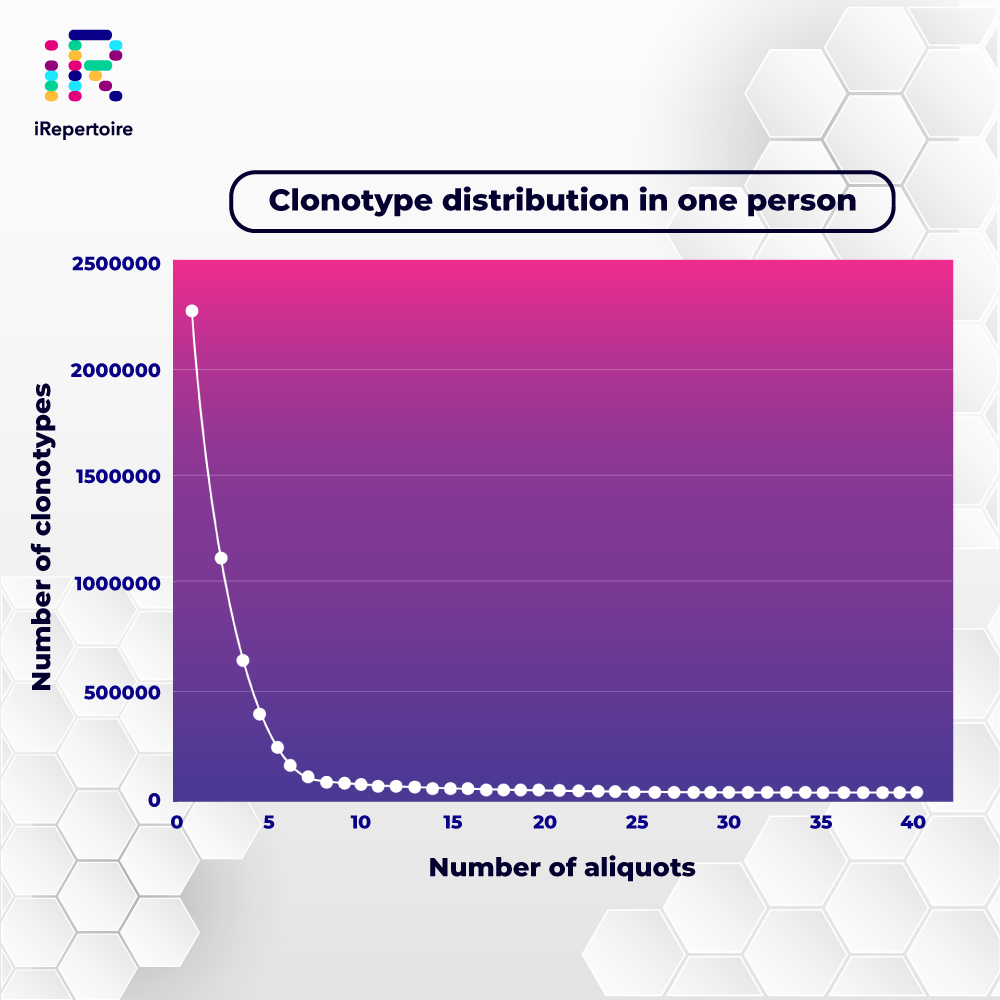
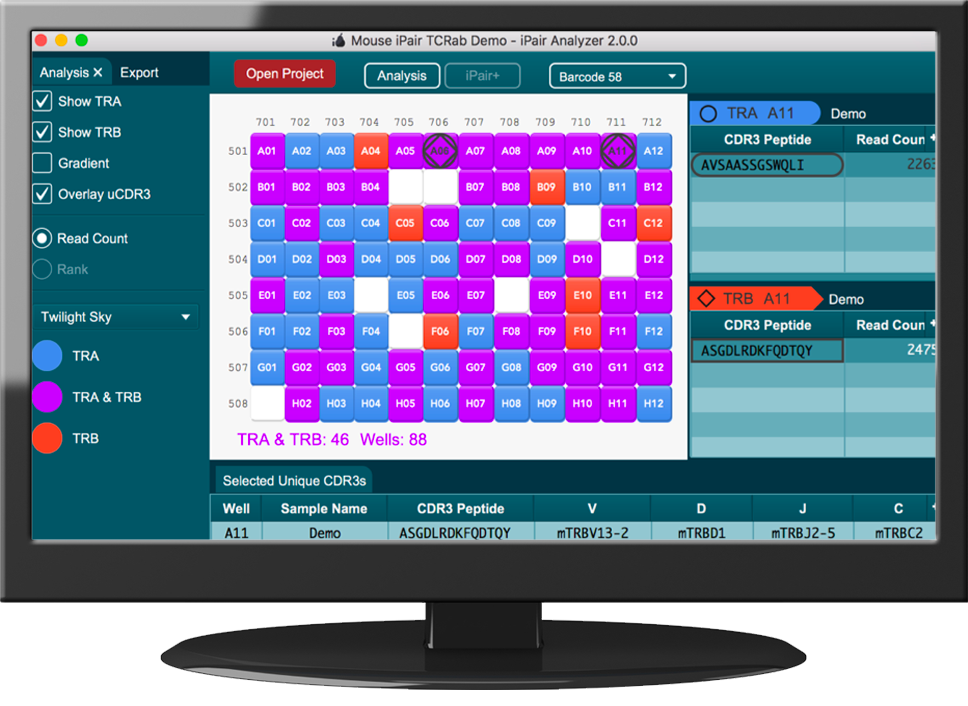
Immunology meets NGS: iRepertoire's Technology
Immunology meets NGS: iRepertoire's Services
Project planning, sample preparation and submission
White papers, case studies, and eBooks
Product manuals
Guides