Technology
Inspiring clinically-actionable discoveries in immune biology
Sequence the full variable region of all 7 chains for single cells or bulk repertoires
An immune repertoire is the sum of functionally diverse T and B cells in a body at any given moment. Sampling
the immune repertoire provides both a snapshot and a historical record of immune function, enabling an advanced understanding of disease and treatment response.
Over the last 10 years, iRepertoire has developed the gold standard technologies for immune repertoire sequencing and continues to innovate new solutions for profiling the adaptome. Our core technologies provide the ability to amplify all of the expressed V(D)Js in B or T cells, and we offer the only technology that can measure all 7 immune chains in a single reaction. iRepertoire is committed to the creation of cutting-edge technology that adapts to the
ever-changing immune sequencing landscape.
arm-PCR
Amplicon Rescue Multiplex PCR
arm-PCR inclusively amplifies all of the expressed V(D)Js in B or T cells for cost-efficient insights into the expressed immune repertoire. arm-PCR is ideal for capturing rare cell types, tetramer analysis, and single cell applications even for low volume and difficult to handle sample types.
dam-PCR
Dimer avoided multiplex PCR
dam-PCR enables highly quantitative amplification of any combination of TCR and BCR chains (alpha, beta, delta, gamma, IgH, and kappa/lambda) in a single reaction. dam-PCR is ideal for isotype and clonotype frequency analysis, hyper-mutation status, and FFPE-damaged samples.
Streamlined single cell immune sequencing for paired chain and phenotypic analysis
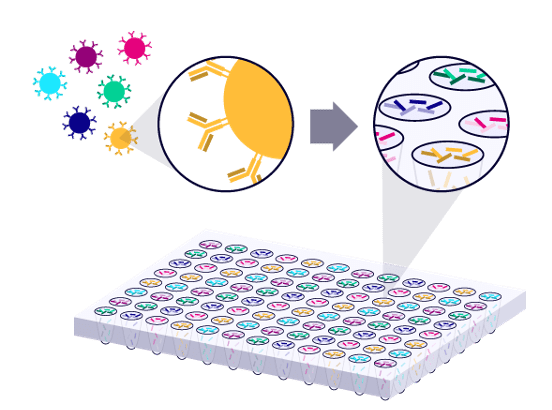
Our iPair single cell immune sequencing technology is the first commercially available service specifically designed for capturing physically paired variable regions in T cells and B cells. Our iPair+ single cell immune phenotyping technology allows for co-expression analysis of over 100 phenotypic genes.
iPair Applications
- Paired chain analysis
- CAR-T development
- Lymphocyte tracking
- Antibody reverse engineering
- Rare cell analysis
Enabling flexible inputs, more insights, and efficiency
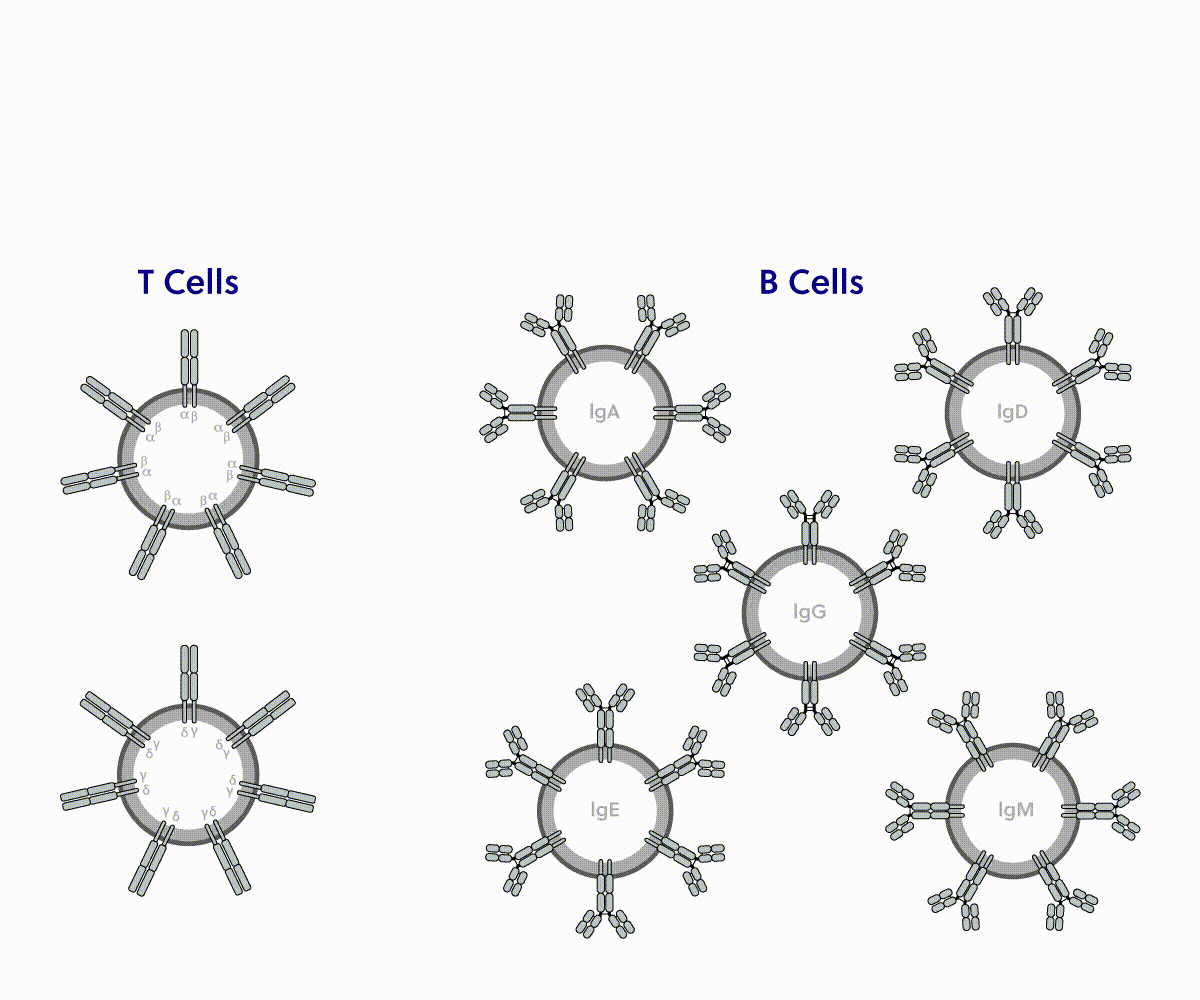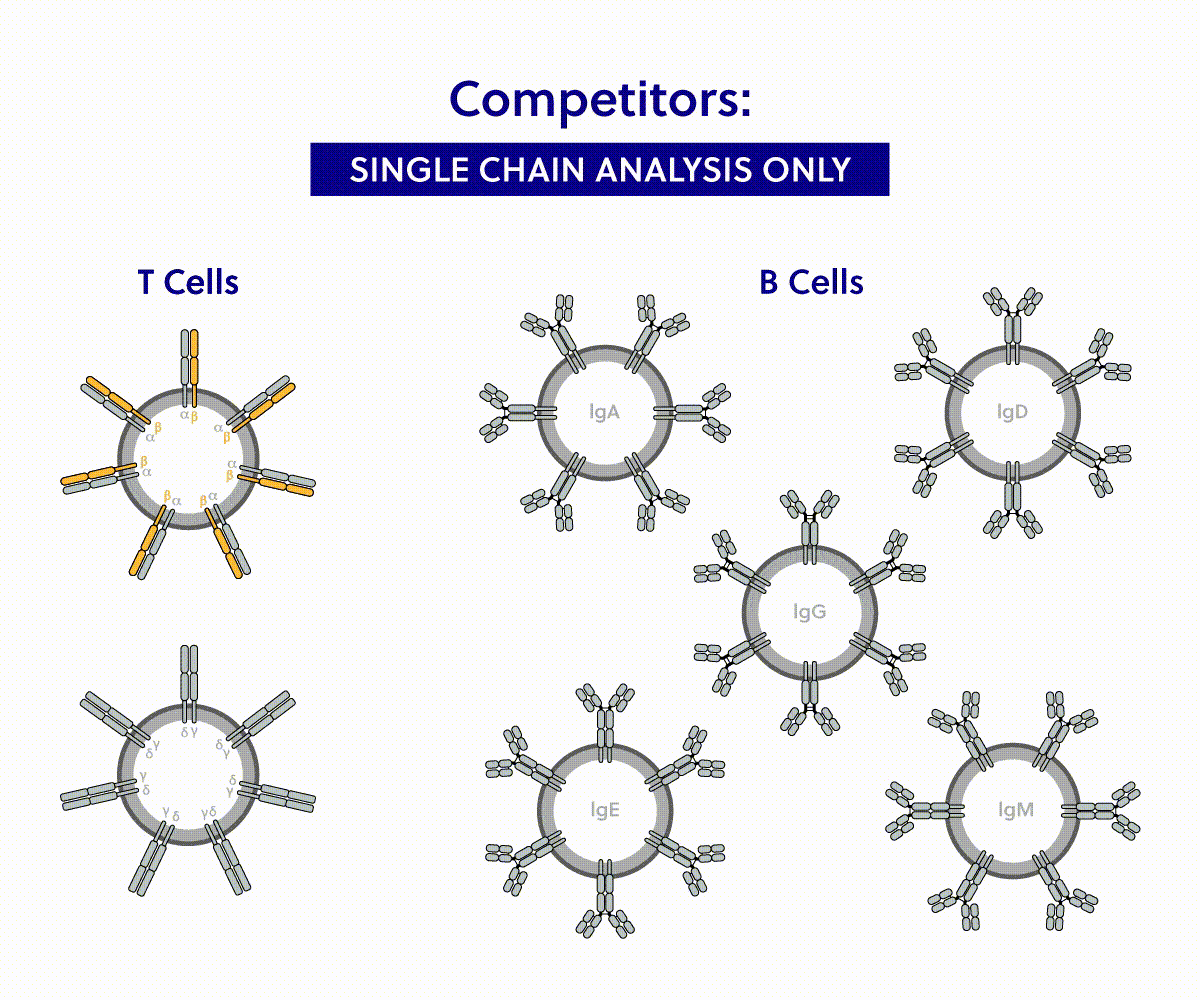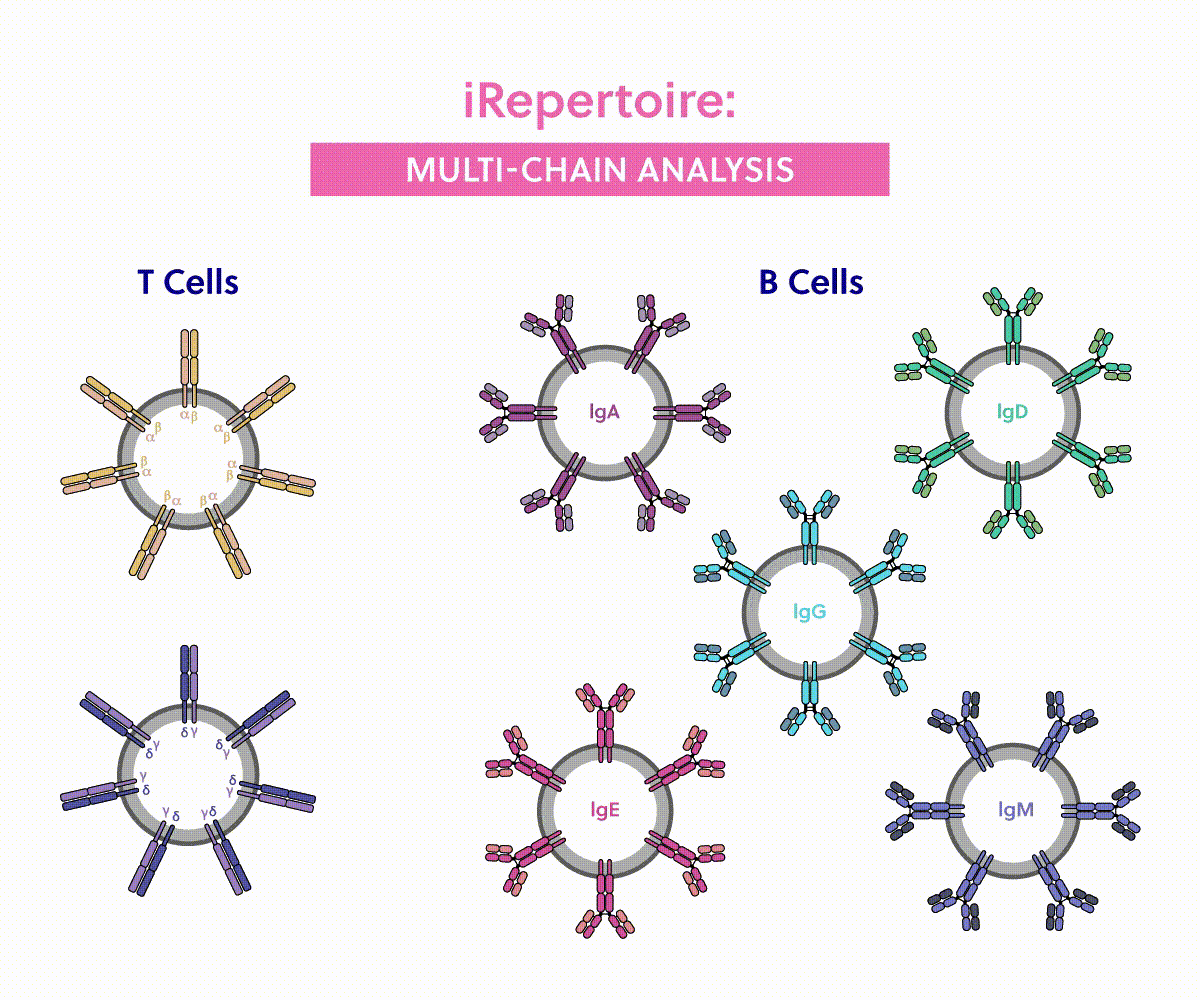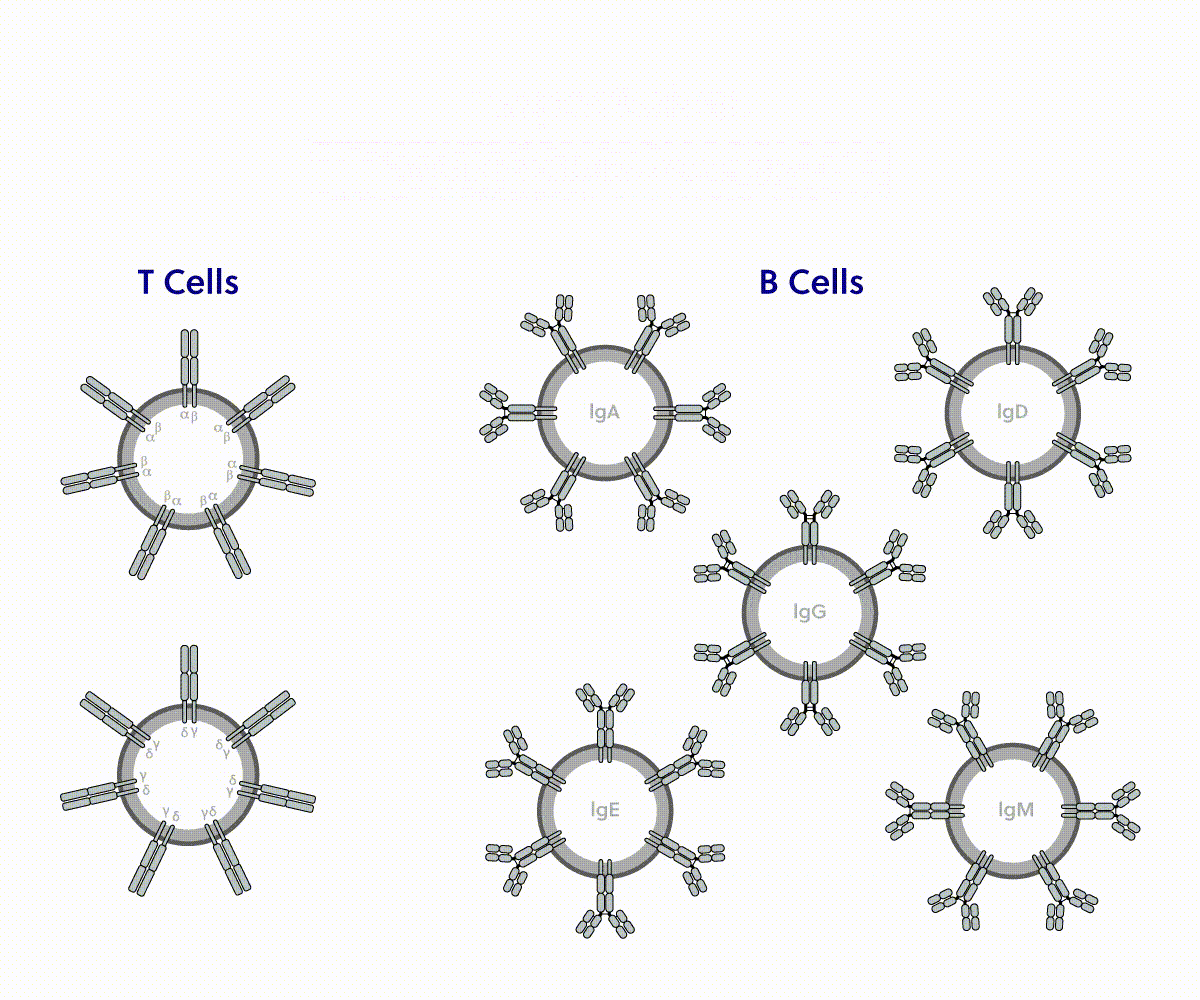
iRepertoire immune sequencing | Competitor analysis
| Coverage | |
|---|---|
| iRepertoire | |
| Technology | arm-PCR and dam-PCR |
| BULK SEQUENCING AVAILABLE? | Yes |
| SINGLE CELL SEQUENCING AVAILABLE? | Yes |
| HUMAN CDR | CDR1, CDR2, & CDR3 |
| MOUSE CDR | CDR1, CDR2 |
| IGH | Human & Mouse |
| IGL | Human & Mouse |
| IGK | Human & Mouse |
| TCRB | Human & Mouse |
| TCRA | Human & Mouse |
| TCRD | Human & Mouse |
| TCRG | Human & Mouse |
| BCR ISOTYPE | Yes |
| UMI AVAILABLE? | Yes |
| INPUT (RNA/DNA) | RNA or DNA |
| PRICE | $ |
| Competitor A | |
| Technology | multiplex-PCR |
| BULK SEQUENCING AVAILABLE? | Yes |
| SINGLE CELL SEQUENCING AVAILABLE? | No |
| HUMAN CDR | CDR3 only |
| MOUSE CDR | CDR3 only |
| IGH | Human & Mouse |
| IGL | Not Covered |
| IGK | Not Covered |
| TCRB | Human & Mouse |
| TCRA | Human |
| TCRD | Human |
| TCRG | Human |
| BCR ISOTYPE | No |
| UMI AVAILABLE? | No |
| INPUT (RNA/DNA) | DNA |
| PRICE | $$ |
| Competitor B | |
| Technology | template switch |
| BULK SEQUENCING AVAILABLE? | Yes (10K cell max) |
| SINGLE CELL SEQUENCING AVAILABLE? | Yes |
| HUMAN CDR | CDR1, CDR2, & CDR3 |
| MOUSE CDR | CDR1, CDR2, & CDR3 |
| IGH | Human & Mouse |
| IGL | Human & Mouse |
| IGK | Human & Mouse |
| TCRB | Human & Mouse |
| TCRA | Human & Mouse |
| TCRD | Human & Mouse |
| TCRG | Human & Mouse |
| BCR ISOTYPE | Yes |
| UMI AVAILABLE? | Yes |
| INPUT (RNA/DNA) | RNA |
| PRICE | $$$ |